Prostate
Author:
Mikael Häggström [note 1]
Contents
TURP: Gross processing
For fragments obtained from transurethral resection of the prostate (TURP):[1]
- Weigh and measure tissue aggregate in 3 dimensions.
- If weight is less than or equal to 12 gm: Submit entirely in biopsy bags (usually approximately 12 cassettes).
- If weight is more than 12 gm: Submit 12 grams in biopsy bags, and 1 additional cassette per each additional 5 grams.
Prostatectomy: Gross processing
- For orientation, apply ink or make a cut to one half of the specimen, and possibly a different one to the other side.[2]
The transverse slices may be cut into quadrants if exceeding laboratory capacity.
See also: General notes on gross processing
Most common targets
- Prostate adenocarcinoma, including cases of high PSA
Evaluation
Screening method
- Before microscopy, look at each microscopy slide by eye, to plan the microscopy screening so as to not miss peripheral fragments.
- Screen at low power, and switch to high power when encountering glandular structures that can not otherwise be cleared. Look in particular for those surrounding nerves.
- At least if no cancer is seen, also look for inflammation.[notes 1]
Characteristics of adenocarcinoma
- Relatively common and highly specific findings of prostatic adenocarcinoma
- [3]
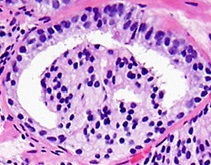
Eccentric nucleoli[3] (pictured example has double and eccentric nucleoli).
- Specific but relatively rare signs of adenocarcinoma
- [notes 2]
On biopsies, look in particular near the tips for perineural invasion, as it is most likely seen by the capsule. Glands adjacent to and indenting nerves is not sufficient as a diagnostic criterion by itself. Glands partially surrounding a nerve is an indication of carcinoma.[4]
- Collagenous micronodules for acinar adenocarcinoma[3]
- Angiolymphatic invasion[3]
- Extraprostatic extension,[3] which in biopsies can be diagnosed when tumor cells are located in fatty tissue.
- Less specific findings
Mitoses: also seen in for example high-grade prostatic intraepithelial neoplasia (HGPIN) and prostate inflammation.[3] Picture shows adenocarcinoma with two mitoses in reactive epithelium.
Intraluminal blue mucin[3] (pictured in acinar adenocarcinoma)
Intraluminal atypical eosinophilic secretions.[3]
Intraluminal crystalloids.[5]
Glomerulations, for acinar adenocarcinoma, consisting of epithelial proliferations into one or more gland lumina, typically a cribriform tuft with a single attachment to the gland wall.[4]
- Prominent nucleoli[3]
- Nuclear enlargement
Precancerous lesions

In case of only less specific findings, consider a Prostatic intraepithelial neoplasia (PIN) or an atypical small acinar proliferation (ASAP).
A PIN is where acini are architecturally benign, but individual cells display atypia. In high-grade PIN (HGPIN), the changes are similar to those of prostate cancer, whereas in low-grade (LGPIN) the changes are milder. Most pathologists do not report the presence of LGPIN.[7]
An ASAP is a lesion that is probably carcinoma but either lacks definitive diagnostic features, or is too small to be certain (see image below).[8] It should not be used for benign lesions that are just unusual looking.[8] In uncertain cases, a diagnosis of adenocarcinoma can be excluded by immunohistochemical detection of basal cells (or confirmed by absence thereof),[3] such as using the PIN-4 cocktail of stains (which consists of P504S, p63 and high-molecular-weight keratins (HMWK) such as CK5 and CK14).
Picture above compares a PIN-4 immunohistochemistry of benign gland (left) and adenocarcinoma (right) using PIN-4. The adenocarcinoma lacks the basal epithelial cells (stained dark brown by p63 and HMWK). Also, in PIN-4 stained samples, adenocarcinoma cells generally display red cytoplasms (stained by AMACR, also known as P504S), while benign glands do not.
Common normal findings
- Infoldings in prostate glands.
- Rectum tissue, by transrectal approach such as biopsies.
Reporting
Example in a normal biopsy:
| (Prostate, right base, needle biopsy:) Benign prostate tissue. |
See Prostate adenocarcinoma article for related reporting.
Notes
- ↑ For a full list of contributors, see article history. Creators of images are attributed at the image description pages, seen by clicking on the images. See Patholines:Authorship for details.
Main page
References
- ↑ Nicole Cipriani. Prostate: TURP. Gross Pathology Manual - The University of Chicago Department of Pathology.
- ↑ Monica Dahlgren, Janne Malina, Anna Måsbäck, Otto Ljungberg. Stora utskärningen. KVAST (Swedish Society of Pathology). Retrieved on 2019-09-26.
- ↑ 3.00 3.01 3.02 3.03 3.04 3.05 3.06 3.07 3.08 3.09 3.10 Cruz, Andrea O.; Santana, Amanda L. S.; Santos, Andréia C.; Athanazio, Daniel A. (2016). "Frequency of the morphological criteria of prostate adenocarcinoma in 387 consecutive prostate needle biopsies: emphasis on the location and number of nucleoli
". Jornal Brasileiro de Patologia e Medicina Laboratorial. doi:. ISSN 1676-2444.
Attribution 4.0 International (CC BY 4.0) license - ↑ 4.0 4.1 4.2 Robert V Rouse MD. Prostatic Adenocarcinoma. Stanford Medical School. Last update 2/2/16
- ↑ Svatek, R S; Karam, J A; Rogers, T E; Shulman, M J; Margulis, V; Benaim, E A (2007). "Intraluminal crystalloids are highly associated with prostatic adenocarcinoma on concurrent biopsy specimens ". Prostate Cancer and Prostatic Diseases 10 (3): 279–282. doi:. ISSN 1365-7852.
- ↑ Image by Mikael Häggström, MD. Reference for features:
- Margaret Sanders, M.B.B.Ch., Murali Varma, M.B.B.S.. High grade prostatic intraepithelial neoplasia (HGPIN). Pathology Outlines. Last author update: 23 February 2021 - ↑ Stanley A Brosman, MD. Precancerous Lesions of the Prostate. Medscape. Updated: Feb 26, 2020
- ↑ 8.0 8.1 . Prostatic Adenocarcinoma - Atypical Small Acinar Proliferation (ASAP). Stanford Medical School. Retrieved on 2020-09-14.
Image sources